International researchers collaborate to investigate transposable element activities and consequences on human pluripotent stem cell transcriptome
27/09/2021
Each cell in a human body contains around 4 billion base pairs, but only 4% is devoted to genes. Nearly 50% of the human genome consists of transposable elements (TEs), self-copying sequences that have extensively colonized the human genome during evolution. TEs consist of several thousand types and include endogenous retroviruses that are molecular fossils of previous infections. The vast majority of TEs are no longer functional, as they have been mutated, yet human cells dutifully copy these TEs to each new generation.
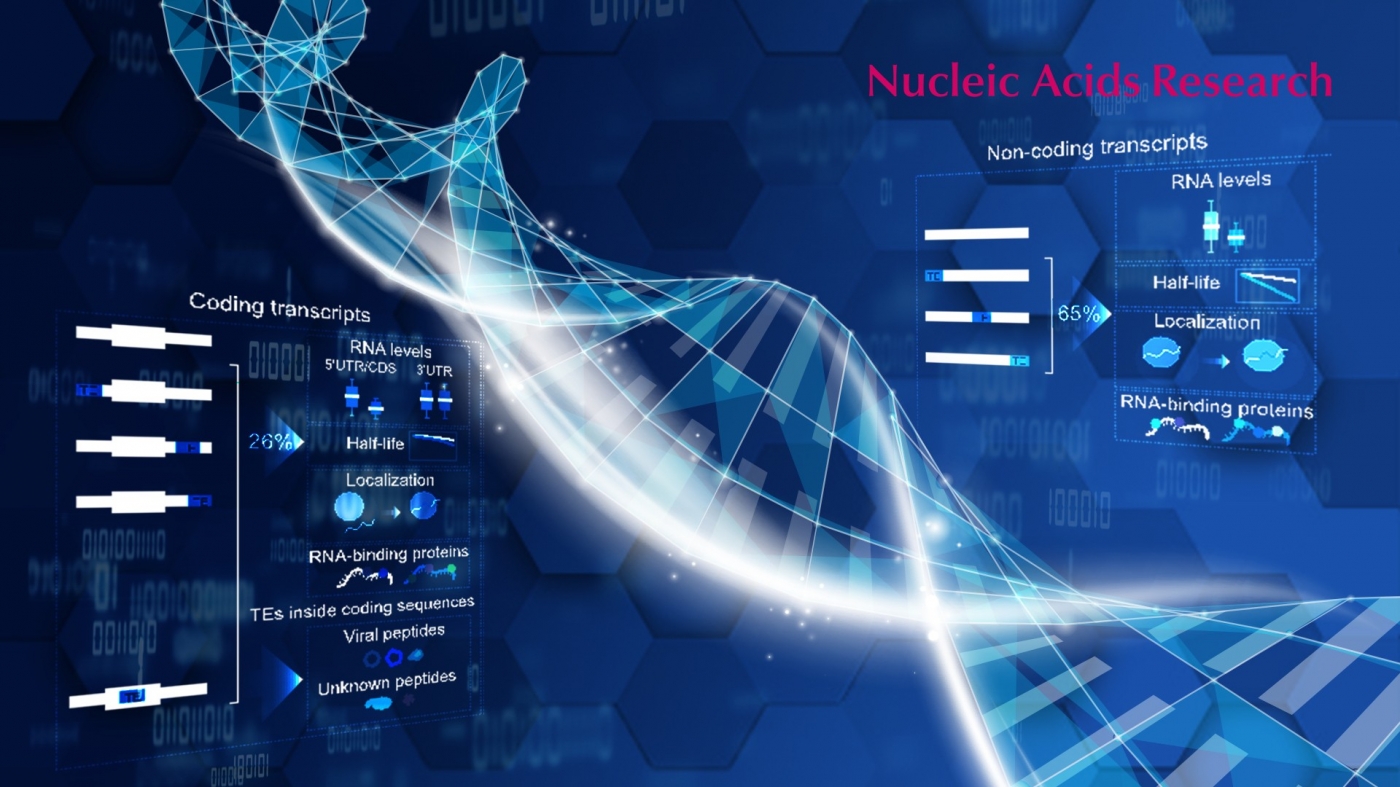
Despite appearing to be junk DNA, TEs have roles in a wide range of cellular activities. TEs can be expressed as RNA and can contribute to the transcriptome of human cells. Many TE-containing transcripts have been identified as critical for cellular function and have roles in human disease. However, due to difficulties in accurately sequencing and analyzing TEs, the full contribution of TE sequences to transcripts has not been explored.
In new research from the Hutchins’ laboratory at the Southern University of Science and Technology (SUSTech), researchers exploit long-read sequencing to assemble full-length transcripts and build a comprehensive map of how TEs contribute to pluripotent stem cell transcripts. This work was published in the leading journal Nucleic Acids Research, entitled, “Transposable element sequence fragments incorporated into coding and noncoding transcripts modulate the transcriptome of human pluripotent stem cells.”
Surprisingly, TE sequences were found in 37% of all transcripts, including 26% of normal protein-coding transcripts. For noncoding transcripts, the majority (65%) had at least one TE sequence fragment. TEs were not randomly distributed in transcripts. In coding transcripts, TEs were mainly found in the 3’UTRs (untranslated regions). However, surprisingly some TEs were directly inside the coding sequence and were translated into fragments of viruses and other proteins of unknown function.
The consequences of TE sequences in a transcript depended on the transcript coding ability and the TE type. TE-containing noncoding transcripts tended to have lower RNA levels, but TEs did not significantly affect the RNA levels of coding transcripts. Generally, TE-containing transcripts were less stable, localized to the nucleus, had non-uniform expression and unique RNA-binding protein (RBP) patterns.
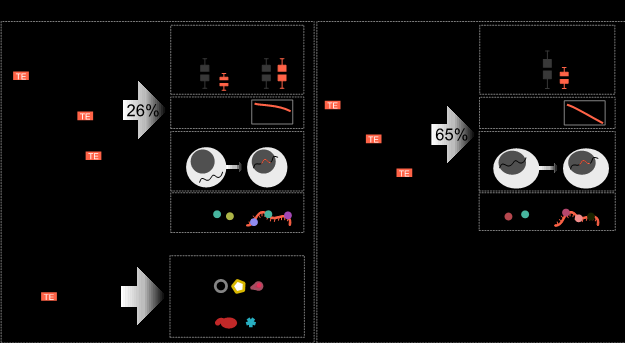
Figure 1. Distribution and function of TEs in different transcripts
Isaac Babarinde, a postdoctoral fellow from SUSTech, is the first author of this paper. Andrew Hutchins of SUSTech is the corresponding author. The work was a collaboration between several research groups, including Miguel Esteban at the Guangzhou Institutes of Biomedicine and Health (GIBH), Ralf Jauch at Hong Kong University, Chen Jiekai at the Bioland Guangdong Provincial Laboratory, Jean-Baptiste Cazier and Jon Frampton from the University of Birmingham, UK, and Tong Guoqing from Shuguang Hospital in Shanghai.
The study was funded by the National Natural Science Foundation of China (NSFC).
Paper link: https://doi.org/10.1093/nar/gkab710
By Yuhao LI & Gang MA
Proofread By Adrian Cremin, Yingying XIA



